Building an XBRL Taxonomy: Widespread Errors and How to Avoid Them

This is a guest post by Katherine Haigh, Quality Assurance Manager at CoreFiling and member of the XBRL International Best Practices Board.
 In this age of digital reporting, hundreds of XBRL taxonomies are published every year. Our analysis at CoreFiling shows that there are a few common mistakes that repeatedly crop up in such taxonomies. These issues can limit their usefulness, slow down implementation and even delay the collection of data. In this blog piece, we will look at what those mistakes are and how to avoid them. They fall into three broad categories: XBRL validity issues, taxonomy package issues and best practice issues.
In this age of digital reporting, hundreds of XBRL taxonomies are published every year. Our analysis at CoreFiling shows that there are a few common mistakes that repeatedly crop up in such taxonomies. These issues can limit their usefulness, slow down implementation and even delay the collection of data. In this blog piece, we will look at what those mistakes are and how to avoid them. They fall into three broad categories: XBRL validity issues, taxonomy package issues and best practice issues.
Editing a taxonomy by hand can often lead to typographical errors which cause the taxonomy to be invalid. One example of this is a difference between the name of a file and a reference to that file, such as a difference in case. Attempting to point to a file called ‘FILE.xml’ with a URI ending in ‘file.xml’ will fail, meaning that the content of the file is not accessible to other files within the taxonomy and the taxonomy becomes invalid.
Other similar errors include duplicate references (due to copy/paste errors) and undeclared namespace prefixes. These can all be avoided by using certified XBRL software to create the taxonomy, or to validate it after creation.
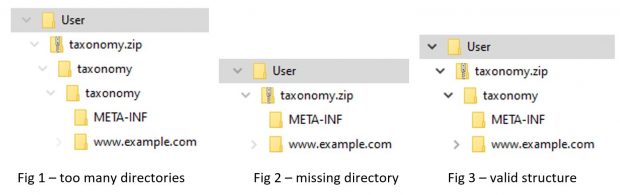
XBRL International recommends that all published taxonomies are provided not only as files on the web, but also as a taxonomy package. The taxonomy package format provides extra information about the taxonomy, such as the entry points used for reporting, in a structured format, and also allows the taxonomy to be quickly installed into compliant software. The most common issue with published taxonomy packages is incorrect directory structure – the package must consist of a zip containing a single top-level directory with the same name as the zip, with taxonomy files inside that directory, but the top-level directory is often omitted or duplicated by taxonomy publishers.
The META-INF directory containing the taxonomy metadata information (taxonomyPackage.xml) and remapping information (catalog.xml) is also a common source of issues – sometimes it is missing or incorrectly placed within the package, and sometimes the files which it should contain are placed elsewhere. Even if the directory contains both a taxonomyPackage.xml and a catalog.xml, errors in their content also crop up, including incorrect ordering of the elements in the taxonomyPackage.xml and remappings to non-XBRL files such as PDFs(!), as well as typographical errors like the ones mentioned above. Once again, these issues can be detected by simply validating the package with certified XBRL software prior to publication. There is also a Practice Profile which explains what a valid package contains.
Best practice issues may not lead to invalid taxonomies, but they can confuse users. Here are some of the top culprits:
| Issue | Why it’s problematic | Example | Solution |
| Shallow remappings | These are likely to overlap with remappings in other packages or those built in to XBRL processing software, and cause conflicts. | Package X depends on Package Y. Both packages include a ‘www.xbrl.org’ directory. Package X uses a file located at www.xbrl.org/files/a/file_a.xsd and package Y uses a file located at www.xbrl.org/files/b/file_b.xsd. Both packages remap ‘http://www.xbrl.org’ to ‘../www.xbrl.org’. XBRL processors don’t know which remapping to use. | Make remappings deep enough to be unique (e.g. Package X should only remap ‘www.xbrl.org/files/a’ and Package Y should only remap ‘www.xbrl.org/files/b’) |
| References to outdated supporting packages, specifications or registries. | Older versions may contain bugs or not support all of the latest taxonomy features. | Use of old namespace xmlns=” http://xbrl.org/PR/2017-02-08/extensible-enumerations-1.1″ which is for an obsolete version of the Extensible Enumerations specification. | Always refer to the latest version of supporting taxonomies, e.g. from https://specifications.xbrl.org/spec-group-index-registries.html |
| Inclusion of non-XBRL files | This makes the taxonomy larger than it needs to be, which can increase memory requirements for processors. | Word/Excel/PDF documents such as release notes and data point model documentation, or copies of files from another taxonomy which could be added to the processor separately. | Provide supporting documentation in a separate ZIP. Don’t copy files from other taxonomies – refer to them at their normative location instead. |
| Lack of backwards compatibility | This can make it difficult for processors to work with multiple versions of the taxonomy. | Version 2.0 of a taxonomy contains some files which are also in version 1.0, and some users need to report using both versions. | Where multiple versions of a taxonomy are valid simultaneously, ensure that their contents do not overlap. |
Hopefully, by following these tips, taxonomy authors can ensure that their taxonomies are consistently high quality every time, ensuring interoperability for everyone’s benefit.